Do you want to find 'thesis molecular docking'? You can find your answers here.
Table of contents
- Thesis molecular docking in 2021
- Protein-ligand interaction ppt
- Protein-ligand interaction pdf
- Docking meaning
- Molecular docking phd thesis
- What is a ligand
- Screening for drugs using molecular docking doctoral thesis
- Thesis molecular docking 08
Thesis molecular docking in 2021
 This image shows thesis molecular docking.
This image shows thesis molecular docking.
Protein-ligand interaction ppt
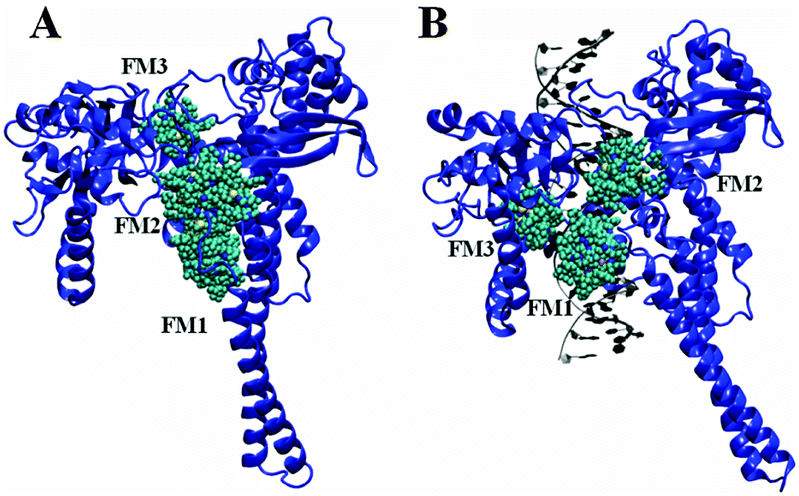 This picture illustrates Protein-ligand interaction ppt.
This picture illustrates Protein-ligand interaction ppt.
Protein-ligand interaction pdf
 This image representes Protein-ligand interaction pdf.
This image representes Protein-ligand interaction pdf.
Docking meaning
 This picture shows Docking meaning.
This picture shows Docking meaning.
Molecular docking phd thesis
 This image shows Molecular docking phd thesis.
This image shows Molecular docking phd thesis.
What is a ligand
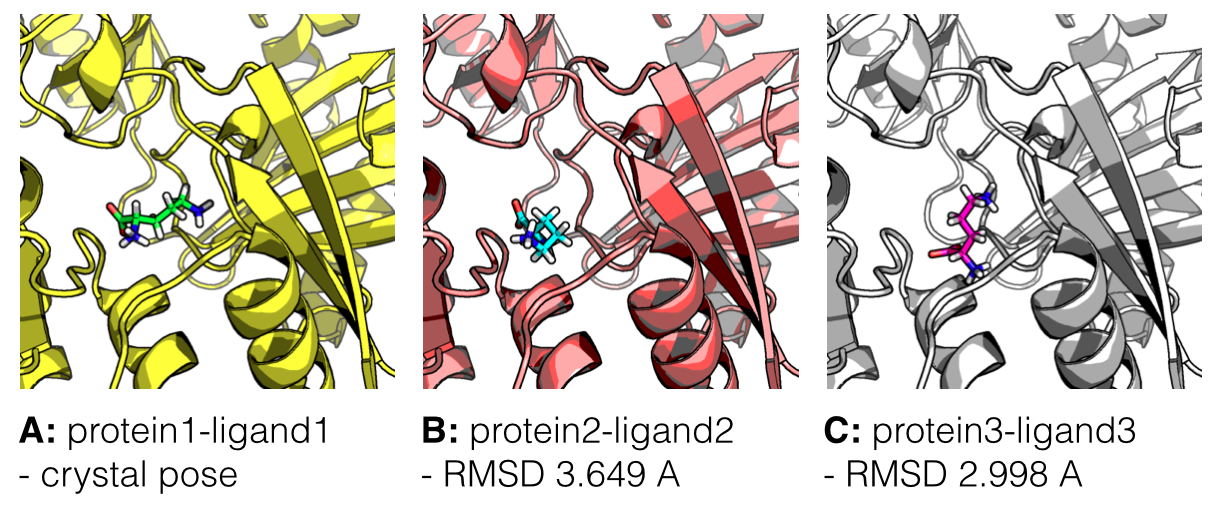 This picture shows What is a ligand.
This picture shows What is a ligand.
Screening for drugs using molecular docking doctoral thesis
 This picture demonstrates Screening for drugs using molecular docking doctoral thesis.
This picture demonstrates Screening for drugs using molecular docking doctoral thesis.
Thesis molecular docking 08
 This image representes Thesis molecular docking 08.
This image representes Thesis molecular docking 08.
How is blind docking used in protein detection?
In the absence of knowledge about the binding sites, cavity detection programs or online servers, e.g. GRID[20, 21], POCKET [22], SurfNet [23, 24], PASS [25] and MMC [26] can be utilized to identify putative active sites within proteins. Docking without any assumption about the binding site is called blind docking.
How is molecular docking used in molecular biology?
The molecular docking approach can be used to model the interaction between a small molecule and a protein at the atomic level, which allow us to characterize the behavior of small molecules in the binding site of target proteins as well as to elucidate fundamental biochemical processes [19].
Why is it important to know binding site before docking?
Knowing the location of the binding site before docking processes significantly increases the docking efficiency. In many cases, the binding site is indeed known before docking ligands into it.
What are the steps in the docking process?
The docking process involves two basic steps: prediction of the ligand conformation as well as its position and orientation within these sites (usually referred to as pose) and assessment of the binding affinity. These two steps are related to sampling methods and scoring schemes, respectively, which will be discussed in the theory section.
Last Update: Oct 2021